Our Research
Unlike most other solid cancers, pancreatic cancer has been among one of the deadliest in decades. The goal of our lab is to fundamentally understand how cancer progresses within the unique microenvironment it creates. Specifically, we are interested in the hypoxic feature in tumor and how it enforces a selective pressure to generate the fittest or most aggressive states. We implement one of the most sophisticated, genetically engineered mouse (GEM) models that allow us to precisely recapitulate the molecular events that drive disease progression in humans. Cancer of these GEM models develop in its natural contexts and have gone through very similar checkpoints as in human disease. Therefore, it is an invaluable tool for the discovery of novel mechanisms of pancreatic cancer progression.
Our first research program is designed to understand how hypoxia drives pancreatic cancer progression. We will use the CRISPR/Cas9 method to identify genes that play essential roles in the survival of cancer cells under the hypoxic condition as well as the adoption of a metastatic phenotype. Combined with both transcriptomic and chromatin accessibility information, we can start to build a comprehensive picture about how cancer cells cope with intratumoral hypoxia and become increasingly metastatic.
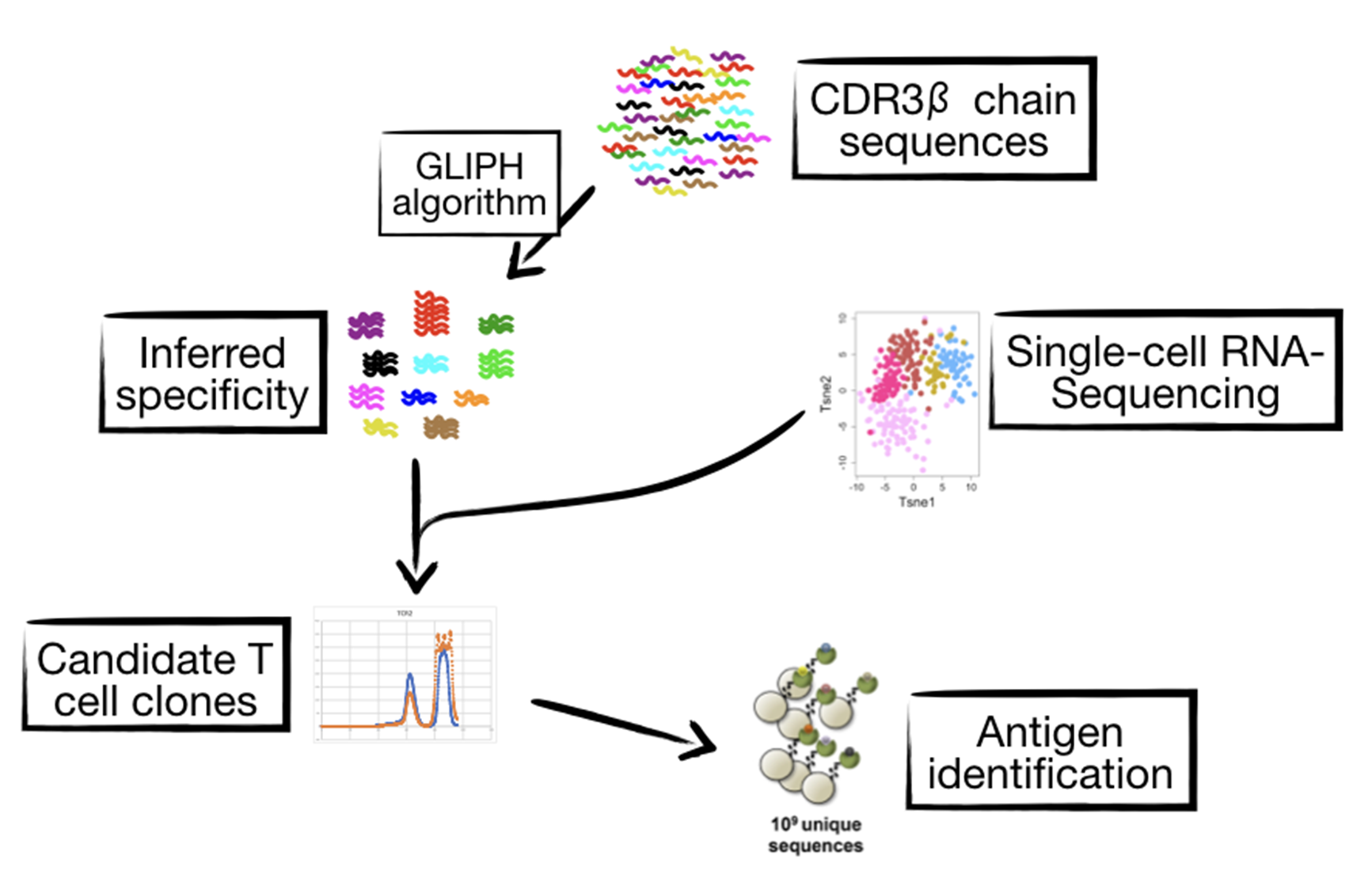
In the light of the recent success in immunotherapy against cancer, we seek to understand why a sizable proportion of patients don’t respond to the immune checkpoint blockade. The basic assumption is that, through immune checkpoint blockade, the rejuvenated adaptive immunity must recognize tumor-specific antigens for the intervention to work. However, very little is known about the antigen specificities of tumor-infiltrating T cells. A better understanding of the specificity landscape of tumor-infiltrating T cells can help us improve the current therapeutic strategies with immune checkpoint inhibitors.
Our second research program starts with massive profiling of the repertoires of tumor-infiltrating T cells in various solid cancer types. Our inhouse bioinformatic tools will prioritize candidate T cell clones that are inferred to be specific to potential tumor antigens with likely human leukocyte antigen (HLA) restriction. We will use yeast libraries displaying a given HLA loaded with up to 10^8-10^9 diverse peptides in order to initially identify mimotopes (mimetics + epitopes) that resemble the cognate antigens for the candidate T cell clones. The superior diversity of the yeast display libraries allows us to identify multiple mimotopes, some of which can potentially map to multiple, cross-reacted antigens. This effort will not only help us catalog a comprehensive T cell specificity landscape in cancer, but also lead to the identification of novel, shared tumor antigens.